The study of complex microbial ecosystems using traditional microbiological methods must first be carried out for the isolation and culture of microorganisms. Due to the complexity of the soil microbial ecosystem and the limitations of culture conditions, the microorganisms that can be cultured in the laboratory are less than 1% of the total number of soil microorganisms . A considerable number of microorganisms are not recognized because they cannot be cultured [1] .
The extraction and purification of soil microbial DNA is a time-consuming and cumbersome process. Many scholars have been exploring faster, more efficient and low-cost methods for soil DNA extraction. Since the mid-1980s, microbiologists have begun to study soil microbes directly using culture-free molecular microbial ecology [2] , which must extract DNA from soil microbial communities. Volossiouk [3] used liquid nitrogen grinding and SDS (sodium dodecyl sulfate) to decompose microbial cells to extract soil DNA, and Tsai [4] used lysozyme and SDS to extract soil DNA, and the DNA extracted by these methods. More impurities, seriously affect the results of subsequent research [5-6] . Later, Bourrain et al [7] extracted the DNA of the microbial community from compost in activated sludge and LaMontagne et al. [8] . However, there is still no rapid, efficient, low cost and high quality method for soil DNA extraction. This article purchased the soil DNA kit commonly used in the market and compared the extraction effects.
The soil samples used for DNA extraction were collected from the barren soil of the Huishan Central Park lawn in Huishan District, Wuxi City (1 to 10 cm topsoil; sample No. 1), the activated sludge on the pond side (sample No. 2), and the rot of the grove. Soil (1 to 10 cm topsoil; sample No. 3).

2.2.1 Grouping
Set up three experimental groups:
( A)O Company (Lot. M****-02);
( B) M Company (Lot. *** 545);
( C) Bioteke (Cat. No.: AU46212).
Soil DNA was extracted strictly according to the kit procedure , and the DNA extraction efficiency of different kits was compared. The extraction amount of DNA extracted by different kits and the feasibility of amplifying 16S rRNA gene were determined by gel electrophoresis and PCR amplification, respectively.
3.1 Comparison of DNA concentration and integrity extracted
Group | sample | A260/A230 | A260/A280 | Sample concentration | unit | Sample type |
Group A | Soil sample 1 | 1.14 | 1.81 | 31.22 | Ng/ul | dDNA |
1.11 | 1.79 | 21.85 | Ng/ul | dDNA | ||
1.17 | 1.8 | 24.31 | Ng/ul | dDNA | ||
Soil sample 2 | 1.46 | 1.84 | 31.62 | Ng/ul | dDNA | |
1.46 | 1.84 | 25.09 | Ng/ul | dDNA | ||
1.53 | 1.86 | 33.71 | Ng/ul | dDNA | ||
Soil sample 3 | 1.73 | 1.84 | 69.37 | Ng/ul | dDNA | |
1.65 | 1.83 | 50.54 | Ng/ul | dDNA | ||
1.47 | 1.81 | 37.03 | Ng/ul | dDNA | ||
Group B | Soil sample 1 | 0.07 | 1.7 | 59.41 | Ng/ul | dDNA |
0.06 | 1.87 | 48.72 | Ng/ul | dDNA | ||
0.06 | 1.83 | 53.26 | Ng/ul | dDNA | ||
Soil sample 2 | 0.07 | 1.94 | 63.65 | Ng/ul | dDNA | |
0.08 | 1.85 | 63.96 | Ng/ul | dDNA | ||
0.07 | 1.93 | 60.32 | Ng/ul | dDNA | ||
Soil sample 3 | 0.09 | 1.87 | 71.07 | Ng/ul | dDNA | |
0.09 | 1.89 | 76.34 | Ng/ul | dDNA | ||
0.08 | 1.9 | 72.47 | Ng/ul | dDNA | ||
Group C | Soil sample 1 | 2.07 | 1.84 | 24.98 | Ng/ul | dDNA |
2.07 | 1.78 | 23.39 | Ng/ul | dDNA | ||
2.36 | 1.83 | 28.05 | Ng/ul | dDNA | ||
Soil sample 2 | 2.31 | 1.87 | 36 | Ng/ul | dDNA | |
2.4 | 1.88 | 37.4 | Ng/ul | dDNA | ||
2.28 | 1.87 | 25.56 | Ng/ul | dDNA | ||
Soil sample 3 | 2.02 | 1.81 | 39.57 | Ng/ul | dDNA | |
2.03 | 1.83 | 40.33 | Ng/ul | dDNA | ||
1.98 | 1.82 | 36.22 | Ng/ul | dDNA |
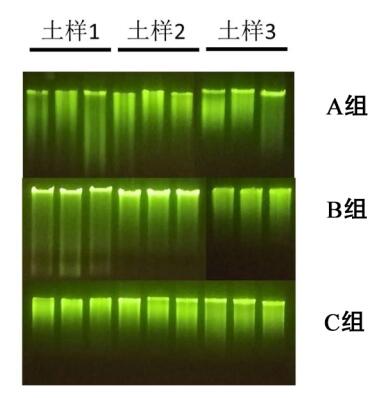
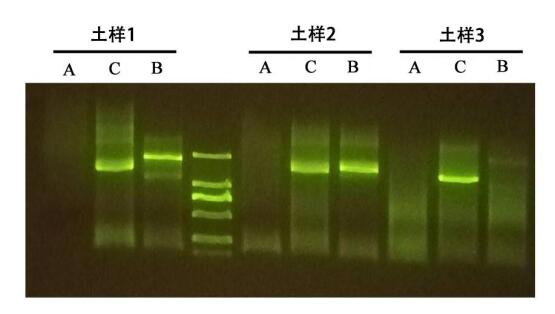
1. The DNA salinity extracted by group A is acceptable, but there are many PCR interference factors, and there is no band in PCR amplification;
2. There are only two kinds of extracted nucleic acids in the three soils of group B that meet the PCR conditions;
3. The soil DNA extracted from the soil extraction kit of group C has less salt residue and high purity. The extracted DNA has low PCR inhibitory factor, and the extracted DNA can be amplified normally, and the strip is single and clear.
Industry Disposable Nitrile Gloves
We, Jiangsu YanFang Medical Technology Co., Ltd, commenced our medical gloves manufacturing in 2020. Currently, we possess a total of 12 high-capacity NBR Glove Dipping Production Lines.
Likewise, we are not only certified with ISO9001, ISO13485 but also fully complied with the essential USFDA, CE Compliances, as well as obtaining relevant accreditation of FDA510K, EN455, and EN374.
Nonetheless, our NBR Examination, Chemotherapy, and Food Grade products are being well established in both US and Europe markets.
We look forward to cooperating and working closely with our valuable customers and stakeholders, who are seeking long-term business relationships in high-quality NBR glove supplies.
Powder-free Nitrile Gloves,nitrile work gloves,safety nitrile gloves,disposable nitrile gloves,industrial gloves
Jiangsu Yanfang Medical Technology Co.,Ltd. , https://www.yanfangchina.com